¶ Description
Apply a R-Based Segmentation model.
¶ Parameters
¶ Parameters tab
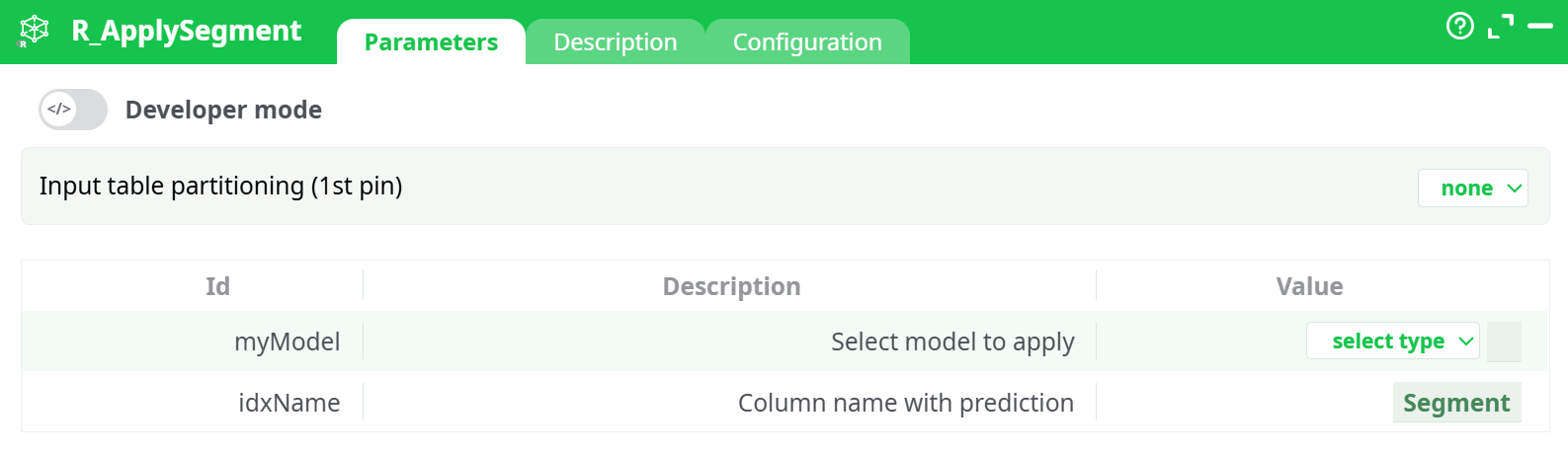
Parameters:
- Input table partitioning (1st pin)
- Select model to apply
- Column name with prediction
¶ Description tab
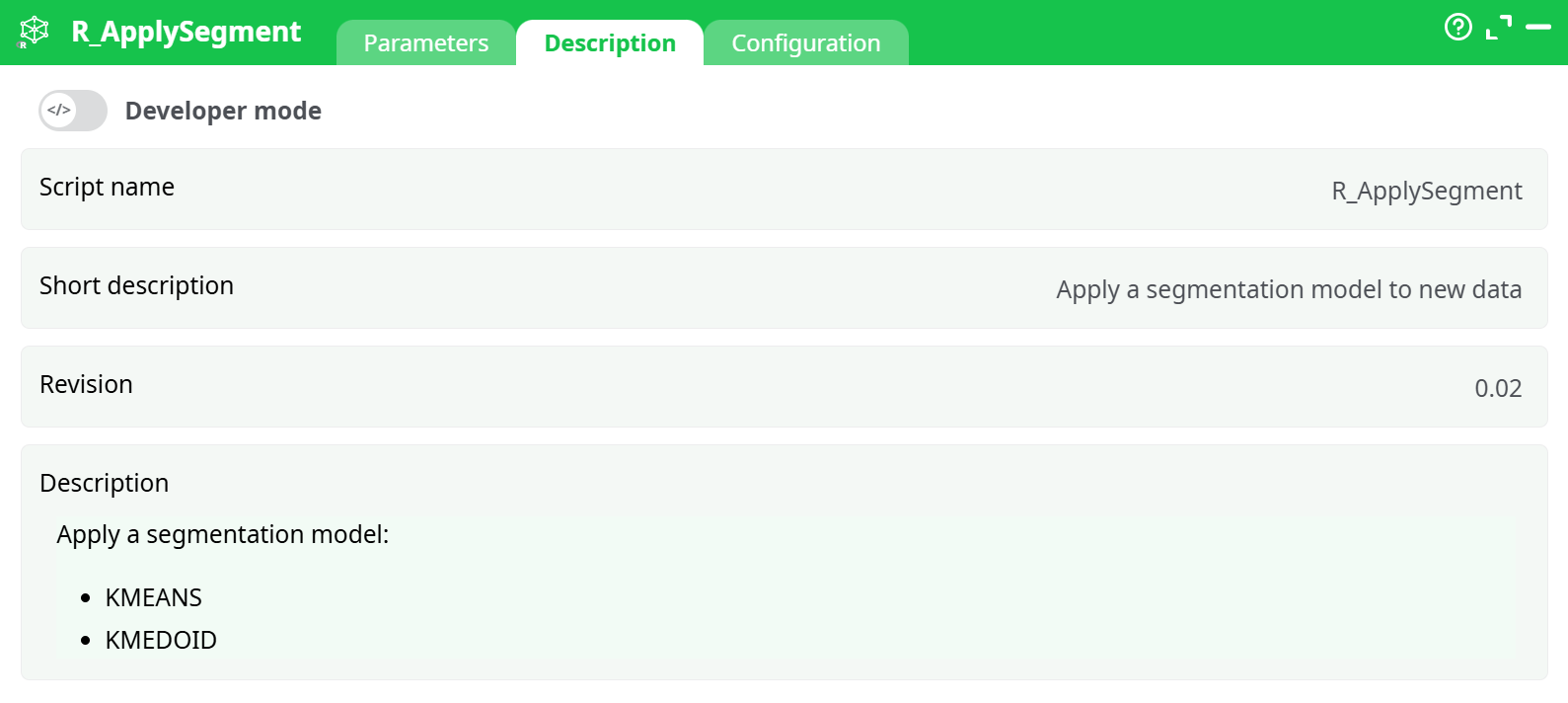
Parameters:
- Script name
- Short description
- Revision
- Decription
¶ Configuration tab
See dedicated page for more information.
¶ About
Use R_ApplySegment to score a table with a previously trained segmentation rModel and append the predicted segment/cluster for every row. Works with very large datasets and all partitioning modes.
¶ What it does
- Loads a segmentation model saved earlier (supported: KMEANS, KMEDOID).
- Checks that the incoming columns match those used at training (same names & compatible types).
- Computes the predicted segment id/label for each input row.
- Returns the original table + one new column containing the segment prediction.
Tip: Train the model with your preferred “R … Segment” training action, then reuse the produced rModel here.
¶ Inputs & outputs
Input (pin 0)
A table with the exact set of features the model expects. You can optionally partition this input (see Input table partitioning).
Output (pin 0)
Same rows and columns as input plus:
Segment(default name) — integer or label of the assigned cluster.
¶ Requirements & assumptions
- Schema match is mandatory: the incoming feature names must be present and compatible with those used when training the segmentation model. Rename or cast upstream if needed.
- No target column needed: this is a pure inference step.
- Partitioning: When using “by column”, ensure that stream is sorted by the chosen column (or enable the Check sorted safety).
¶ Typical workflow
- Train your segmentation model (KMEANS or KMEDOID) with a training action that outputs an rModel file.
- Connect your scoring dataset to R_ApplySegment.
- Select the
myModelrModel from assets (or your preferred source). - (Optional) Set
idxNameto your desired output (e.g.,cluster_id,customer_segment). - (Optional) Choose a partitioning strategy to scale to very large data.
- Run. The output table has all original columns + the segment prediction.
¶ Performance tips
- Large tables: Use fixed number of lines partitioning (size tuned to your RAM).
- Group-scoring: Use by column partitioning (e.g.,
country), and ensure the stream is sorted. - Parallelism: Enable Allow parallel execution when partitions are independent and you have spare CPU.
- I/O throughput: If rows are wide, increase Shared memory size to reduce IPC overhead.
¶ Troubleshooting
- “Unknown column …” / mismatch errors
The scoring table doesn’t match the training schema. Add a Select/Rename/Cast step upstream to align names and types. - All rows get the same segment
Check that feature columns are not constant/missing after joins or filters. Also verify that scaling/encoding upstream matches training-time processing. - Partitioning error (not sorted)
When using by column, either sort upstream or disable the strict check (not recommended). - Performance is slow
Use partitioning + parallel execution; bump Shared memory size; prefer reading the model from assets or @shared storage.
¶ Best practices
- Keep a feature contract: store alongside the rModel a short manifest listing expected columns & types.
- Name the output column clearly (
segment,cluster_id) to simplify downstream grouping and KPIs. - Version your models (e.g.,
seg_kmeans_v3.rModel) and record training metadata.
¶ Example (quick start)
- myModel:
assets:/models/seg_kmeans_v3.rModel - idxName:
cluster_id - Input table partitioning:
fixed number of lines (excluding last)= 250 000 - Configuration: Allow parallel execution = ON, Shared memory size = 8 MB
Result: output table with cluster_id appended.
¶ Notes & compatibility
- Supports segmentation models created by the platform’s R-based trainers (KMEANS/KMEDOID).
- Produces deterministic results given the same model and input (partitioning does not change assignments).
