¶ Description
Greenleaf's Yaysaying control.
¶ Parameters
¶ Parameters tab
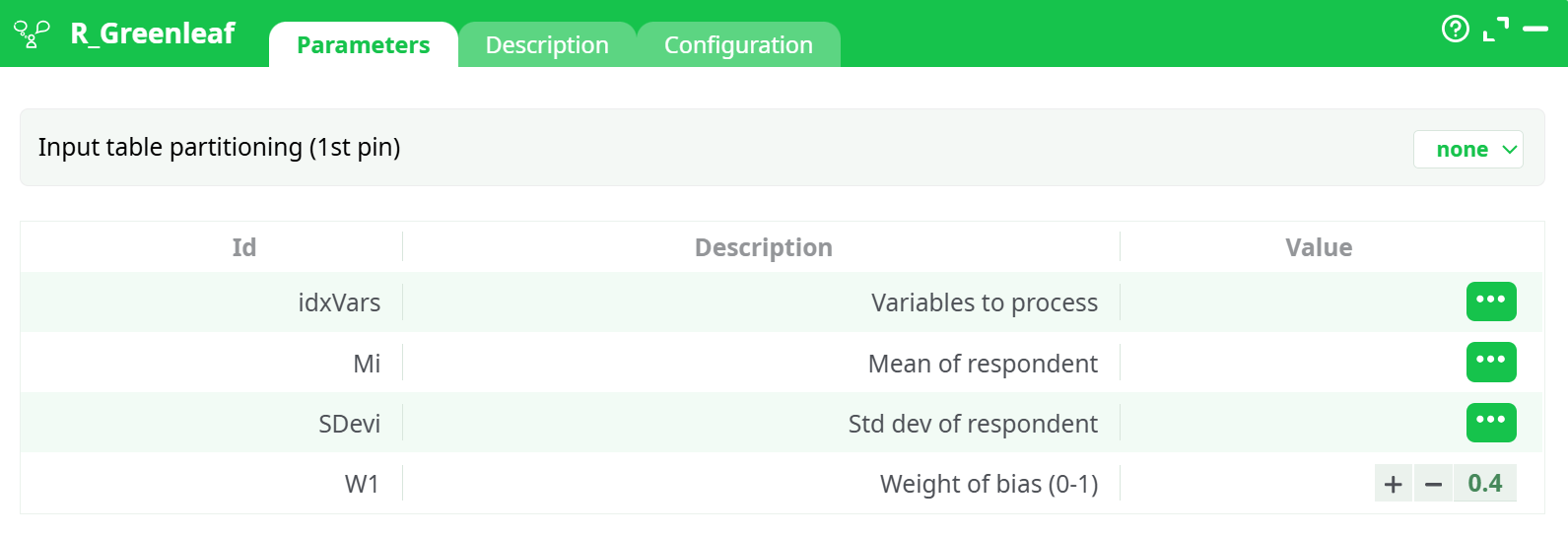
Parameters:
- Partitionnement de la table d'entrée (1ère broche)
- Table entière
- Variables to process
- Mean of respondent
- Std dev of respondent
- Weight of bias (0-1)
¶ Description tab

Parameters:
- Script name
- Short description
- Revision
- Description
¶ Configuration tab
See dedicated page for more information.
¶ About
R_Greenleaf applies Greenleaf’s method to reduce systematic agreement bias in questionnaire responses.
Given each respondent’s mean (Mi) and standard deviation (SDevi) across a set of items, the action outputs adjusted scores (GL_<item>) for the chosen item columns. The strength of the correction is controlled by W1 (0–1).
Control acquiescence (“yaysaying”) bias in Likert-style survey data by adjusting each respondent’s item scores using their personal mean and dispersion. Produces bias-corrected scores for a selected set of items.
¶ Input
¶ Required table (1 row = 1 respondent)
- One numeric column per item to correct (e.g.,
Q1,Q2,Q3) — all on the same Likert scale (e.g., 1–5). Mi— respondent’s mean across the selected items.SDevi— respondent’s sample standard deviation across the selected items.
Accepted formats: CSV, table from upstream action. All selected columns must be numeric.
Minimal example (CSV):
Mi,SDevi,Q1,Q2,Q3
4.000,1.000,5,4,3
3.333,0.577,4,3,3
2.667,1.155,2,2,4
4.000,1.000,3,4,5
1.333,0.577,1,2,1
Tip: If your source file contains only item columns, compute
MiandSDevirow-wise first (e.g., with a “Row Stats/Row Aggregates” action).
¶ Output
- For every selected item column, the action emits a bias-corrected column named
GL_<item>(e.g.,GL_Q1,GL_Q2,GL_Q3) on the original scale. - The original columns are kept unless you remove them downstream.
Example (first 5 rows):
Mi SDevi Q1 Q2 Q3 GL_Q1 GL_Q2 GL_Q3
4.000 1.000 5 4 3 4.76 4 3
3.333 0.577 4 3 3 3.90 3 3
2.667 1.155 2 2 4 1.80 1.80 3.80
4.000 1.000 3 4 5 3.00 4 4.76
1.333 0.577 1 2 1 0.60 2.20 0.60
Interpretation:
GL_*values are the item scores with acquiescence bias reduced using each respondent’sMiandSDevi. Higher W1 → stronger correction; W1 = 0 → no change.
¶ How to Run
-
Prepare input data
Ensure you have columnsMi,SDevi, and the item columns to correct (Q1..Qk). All must be numeric. -
(If reading CSV) Import data
readCSV: delimiter,, decimal.- If your platform does not enforce types, insert ChangeDataType and cast
Mi,SDevi,Q1..Qkto Float/Double.
-
Open R_Greenleaf → Parameters
idxVars→ selectQ1, Q2, Q3(or your item set)Mi→MiSDevi→SDeviW1→0.4(adjust as desired)- Input table partitioning (1st pin) →
none
-
Run
Inspect the Data tab of the action’s output for new columnsGL_Q1..GL_Qk.
Optionally add writeCSV to export results.
¶ Example Pipeline
readCSV → ChangeDataType (cast Mi,SDevi,Q1..Qk to float) → R_Greenleaf → writeCSV
¶ Screenshot Placeholders
-
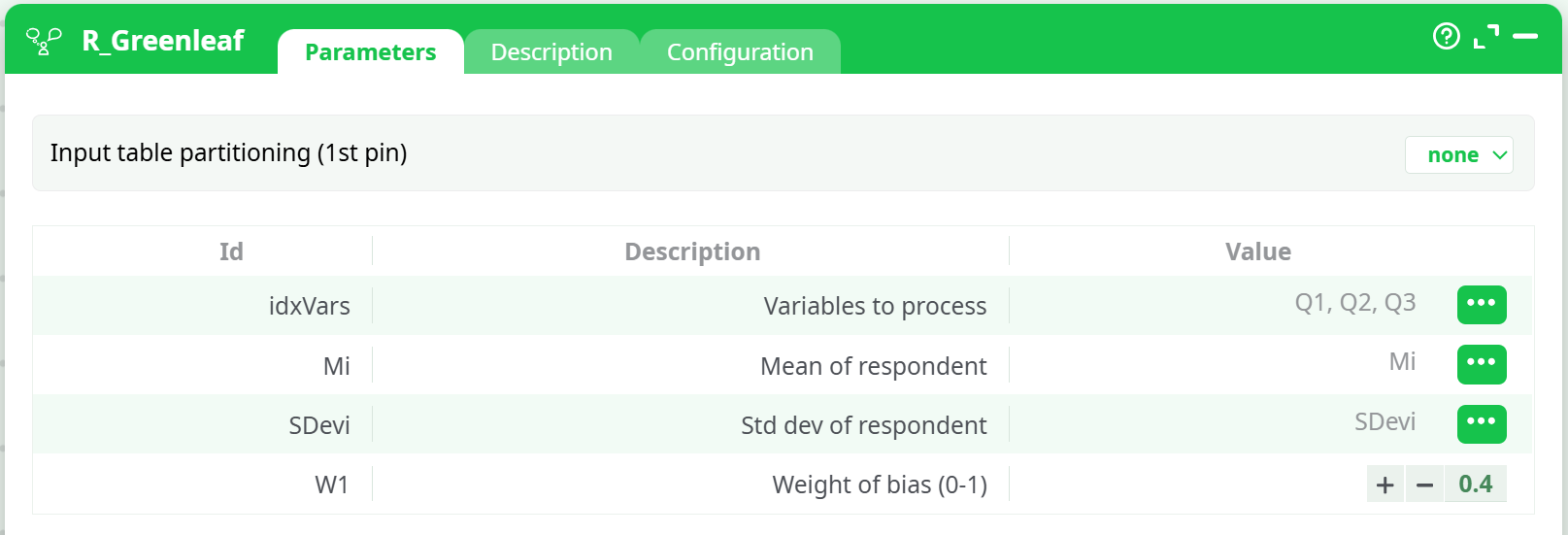
ShowsidxVars=Q1,Q2,Q3,Mi=Mi,SDevi=SDevi,W1=0.4, and partitioning=none. -
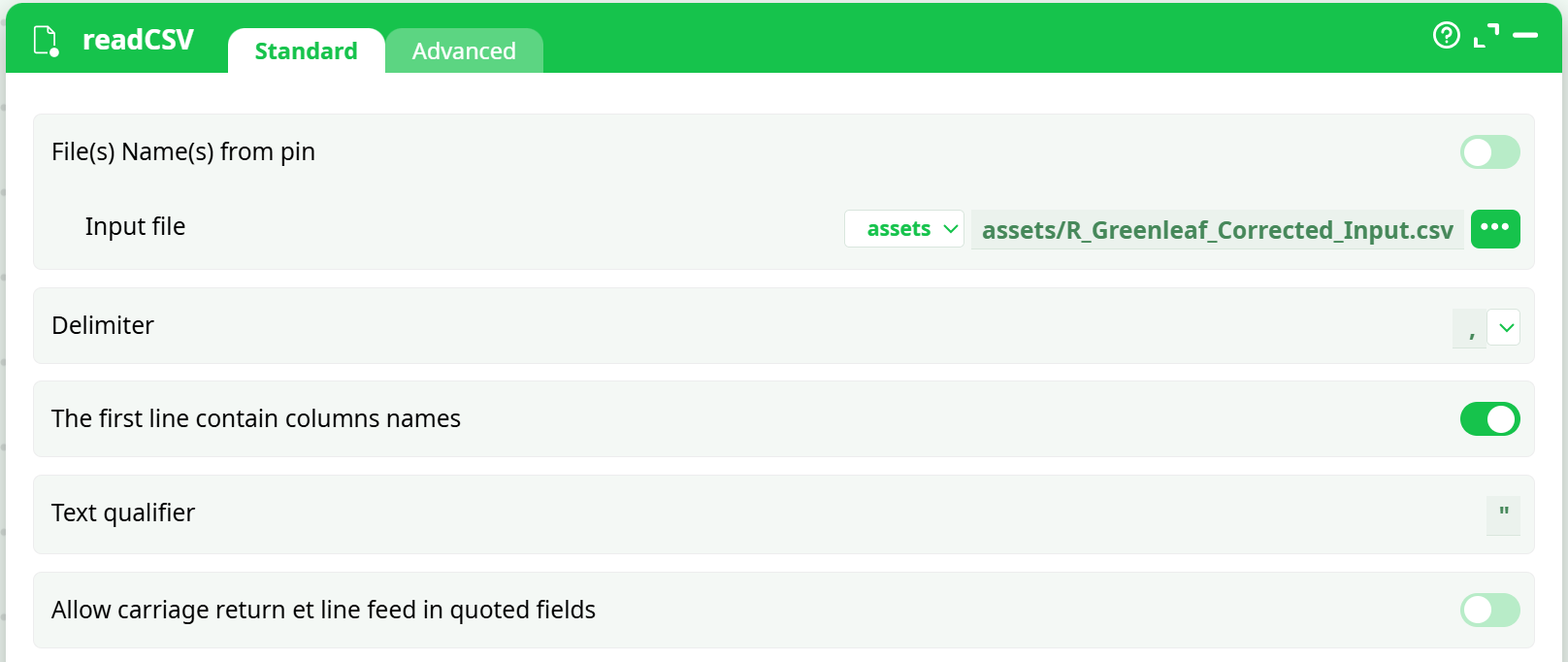
Demonstrates delimiter,, decimal., header row ON. -
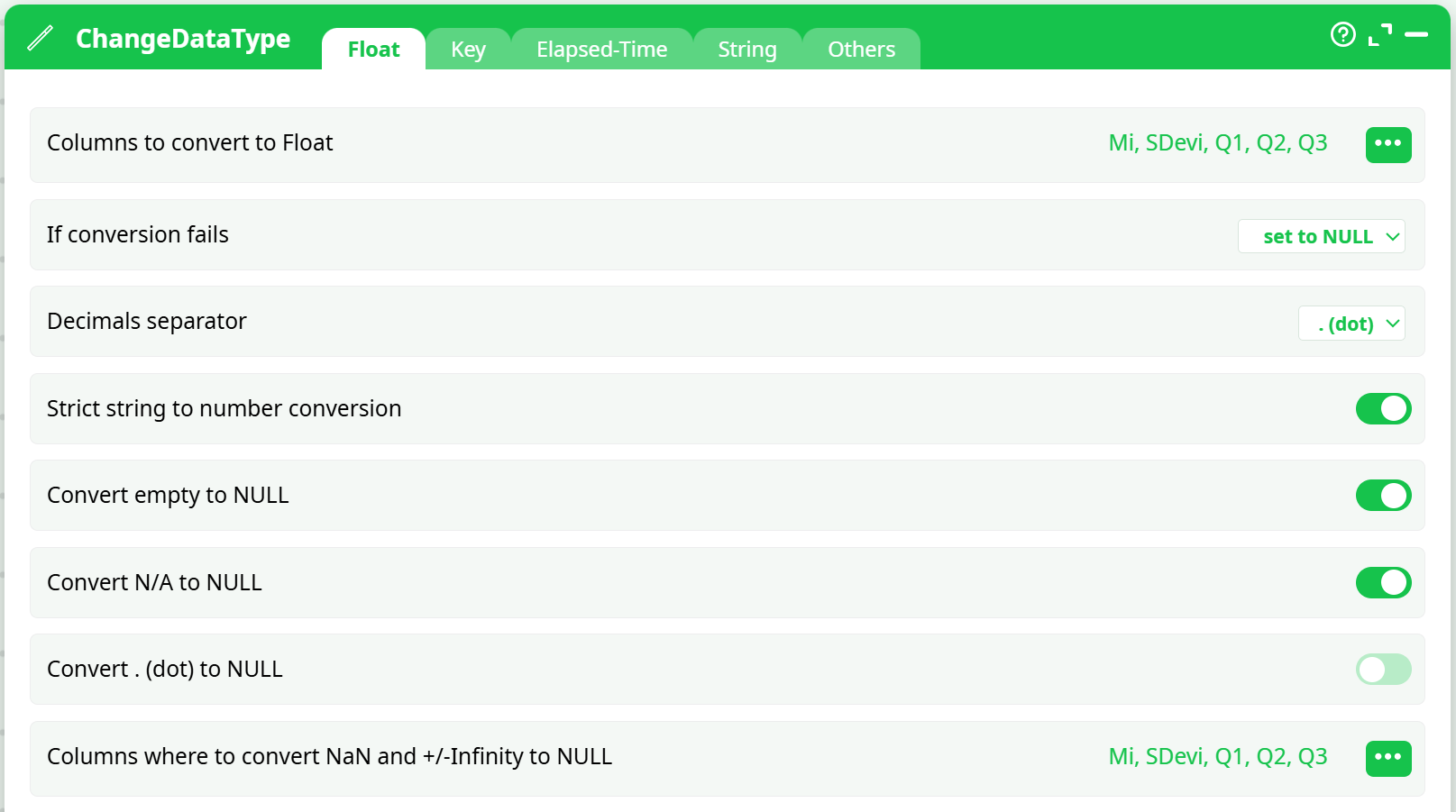
Illustrates casting ofMi,SDevi,Q1,Q2,Q3to Float; strict conversion ON. -
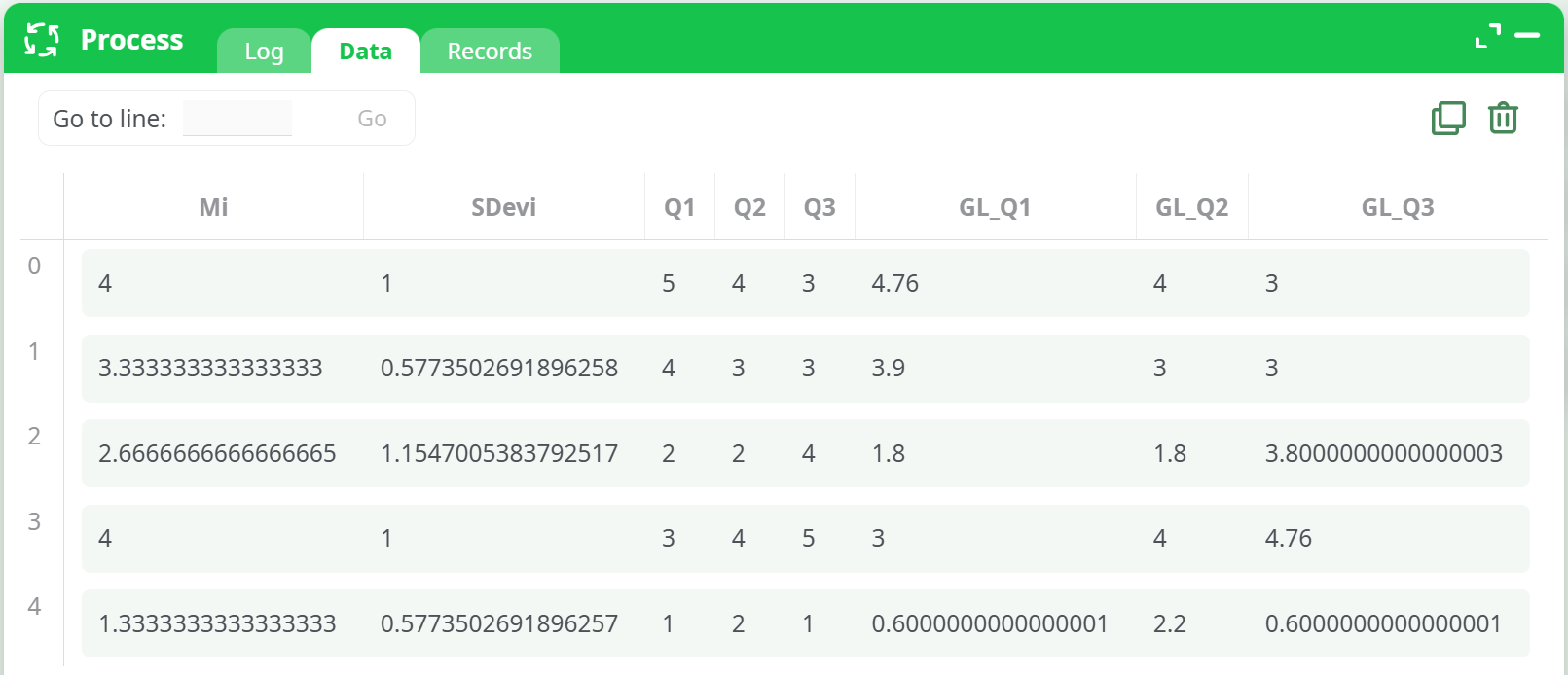
Displays resulting table includingGL_Q1,GL_Q2,GL_Q3. -
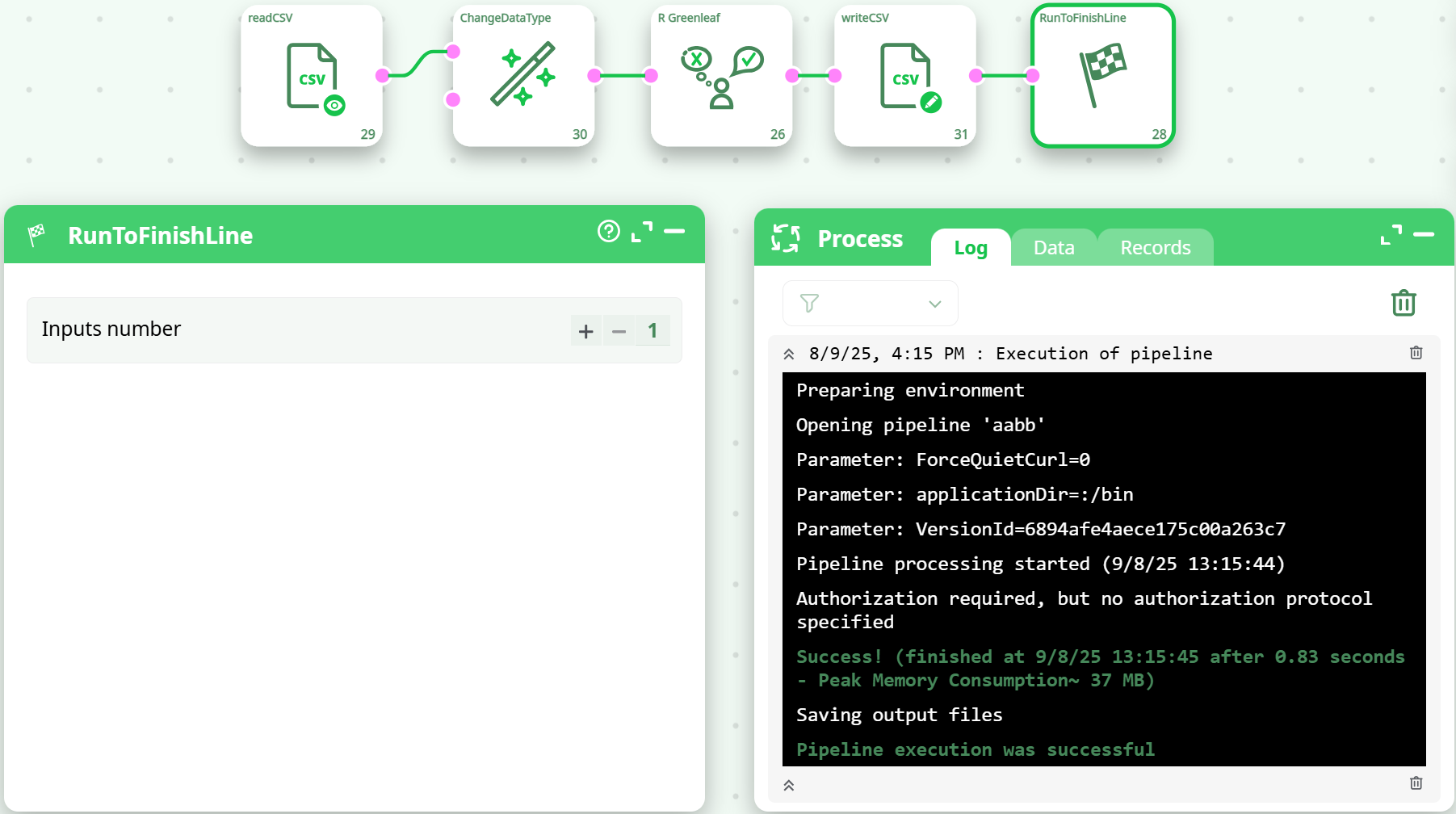
Confirms successful execution and file save.
¶ Notes & Best Practices
- All item columns must be numeric. If you see
non-numeric argument to binary operator, cast types before R_Greenleaf. - Same scale across items. Don’t mix 1–5 with 1–7 items.
- Missing values: If present, consider imputing or let them flow as
NULL; downstream stats should account for them. - Sensitivity (W1): Start at
0.3–0.5. Increase only if diagnostics indicate strong acquiescence bias. - Reproducibility: Save the exact item list used in
idxVarsand the chosenW1.
¶ Appendix — Quick Validation Checklist
-
Mi= row-mean across the selected items -
SDevi= row sample SD across the selected items -
idxVarsincludes only numeric item columns -
W1between 0 and 1 - Output contains
GL_<item>columns on the same scale as inputs
